Below we present possible applications of the database and server that should help researchers understand how topologically interlinked single and multiple chains and macromolecular complexes influence physicochemical properties of proteins. All information deposited in LinkProt also can be used to identify homological proteins with the same topology or identify known existing types of links when new protein complex is determined e.g. based on the EC method.
LinkProt database can be used to understand unexpected thermal stability of pairs of proteins which are interlinked by cysteine bridges (deterministic links). It is well known that disulfide bridges can provide stability to the structure, especially in an extracellular environment, however topological analysis can explain why some intracellular proteins show very high thermal stability. One example deposited in the database is citrate synthetase with PDB code 2IBP from Pyrobaculum aerophilum, a hyperthermophilic archaeon which was discovered by Boutz et al. (2007) [1]. This homodimer possesses two intermolecular disulfide bridges, which close each subunit. These subunits are arranged in such way that they form non-trivial topology of the Hopf link (with positive orientation), as shown in Figure 1. When a minimal surface is spanned on each closed loop (the red and blue surfaces), it is possible to determine position along the chain, which pierces such a surface, as shown in Fig. 1.
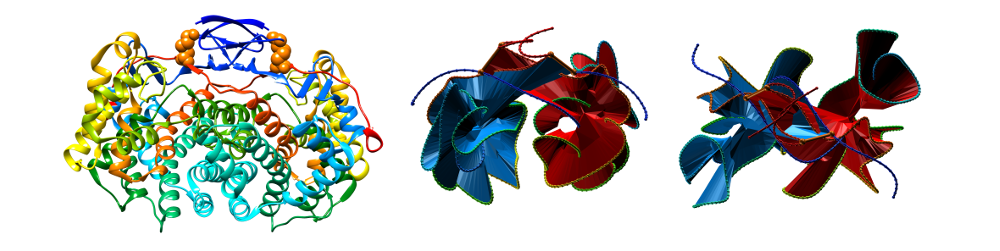
[1] Boutz DR, Cascio D, Whitelegge J, Perry LJ and Yeates TO. Discovery of a thermophilic protein complex stabilized by topologically interlinked chains. J.Mol. Bio., 2007, 368, 1332-1344.
Extraordinary stability due to topologically interlinked chain - capsid.
The LinkProt can also be used to explain (visually) extraordinary stability in molecular complexes, e.g. in the bacteriophage HK97 capsid [1], for which it was shown that this unique property cannot be disassembled to enter an SDS gel [2] without protease treatment [3]. An extraordinary stability of the bacteriophage HK97 capsid is a consequence of its nontrivial topology, i.e. a presence of a link, which is fromed by domains joined to two of its neighbors by ligation of two aminoacids (the side-chain lysine 169 to asparagine 356) [1], as shown in Fig. 2 and 3). To date, a few other structures of capsids with non-trivial topologies were found:
LinkProt can be used to obtain precise information about topology and the presence of links, and to identify subunits (or individual amino acids) that cross the surface spanned on macromolecular rings. The links can be formed as in bacteriophage HK97, via the side-chain joining of lysine 169 to asparagine 356, or by closing neighboring N- and C-terminal. The bacteriophage BPP-1 capsid is composed of three types of links - the Hopf link (one pentameric and one hexameric ring or two hexameric rings), the (one pentameric and two hexameric rings), and the "flower" link (one pentameric and five hexameric rings, as defined by us), when the terminals of domains are closed, as shown in the Fig. 4.
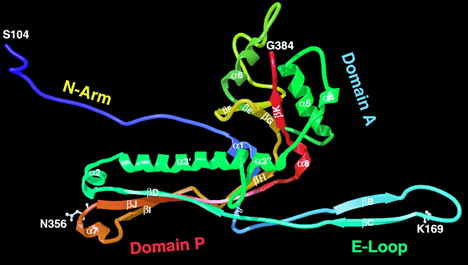
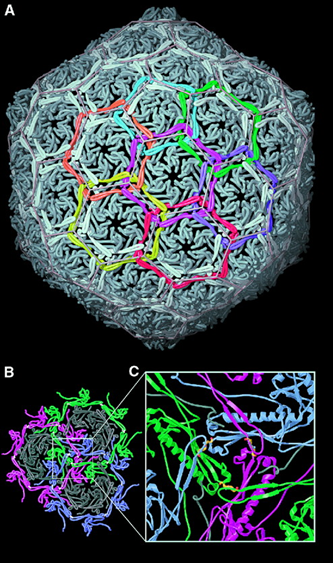
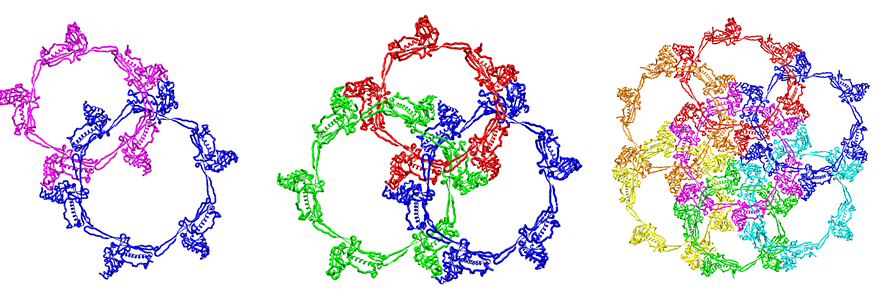
[1] WR Wikoff, L Liljas, RL Duda, H Tsuruta, RW Hendrix, JE. Johnson. Science, 289, 5487, pp. 2129-2133, 2002.
[2] RL Duda, Cell 94, 55, 1998.
[3] MP Popa, TA McKelvey, J Hempel, R W. Hendrix, J. Virol. 65, 3227, 1991.
[4] A Fokine, PG Leiman, MM Shneider, B Ahvazi, KM Boeshans, AC Steven et al. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry, Proc Natl Acad Sci USA, 102, 7163-7168, 2005.
[5] F Akita, KT Chong, H Tanaka, E Yamashita, N Miyazaki, Y. Nakaishi et al.,
The crystal structure of a virus-like particle from the hyperthermophilic archaeon Pyrococcus furiosus provides insight into the evolution of viruses, J Mol Biol, 368, 1469-1483, 2007.
[6] M Sutter, D Boehringer, S Gutmann, S Gunther, D Prangishvili, MJ Loessner et al., Structural basis of enzyme encapsulation into a bacterial nanocompartment, Nat Struct Mol Biol, 15, 939-947, 2008.
[7] X Zhang, H Guo, L Jin, E Czornyj, A Hodes, WH Hui, AW Nieh, JF Miller, ZH Zhou, A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 Å resolution, eLife 2:e01299, 2013.