There are two main options a user can choose to view or analyze data:
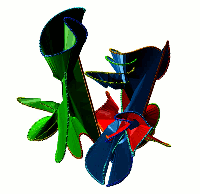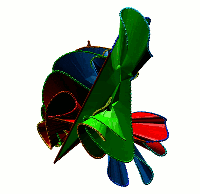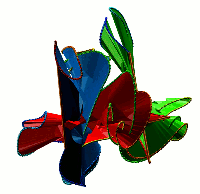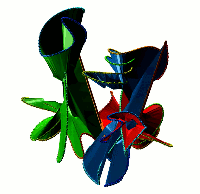
Choosing this option results in the list of all non-trivial, non-artificial linked proteins (either deterministic or probabilistic) stored in the database. There are three ways the database can be browsed (Fig. 1) which can be change upon clicking on one of the small icons in the top-left corner. In the default view the normal list is printed, inculding, for each protein its topological type, way of joining the termini (deterministic or probabilistic), likelihood of the link and the PDB code. The way the termini are connected is described by the colour square - the black square denote deterministic links, while blue square denotes the probabilistic links. For the proteins with gap in the structure additional symbol is visible.
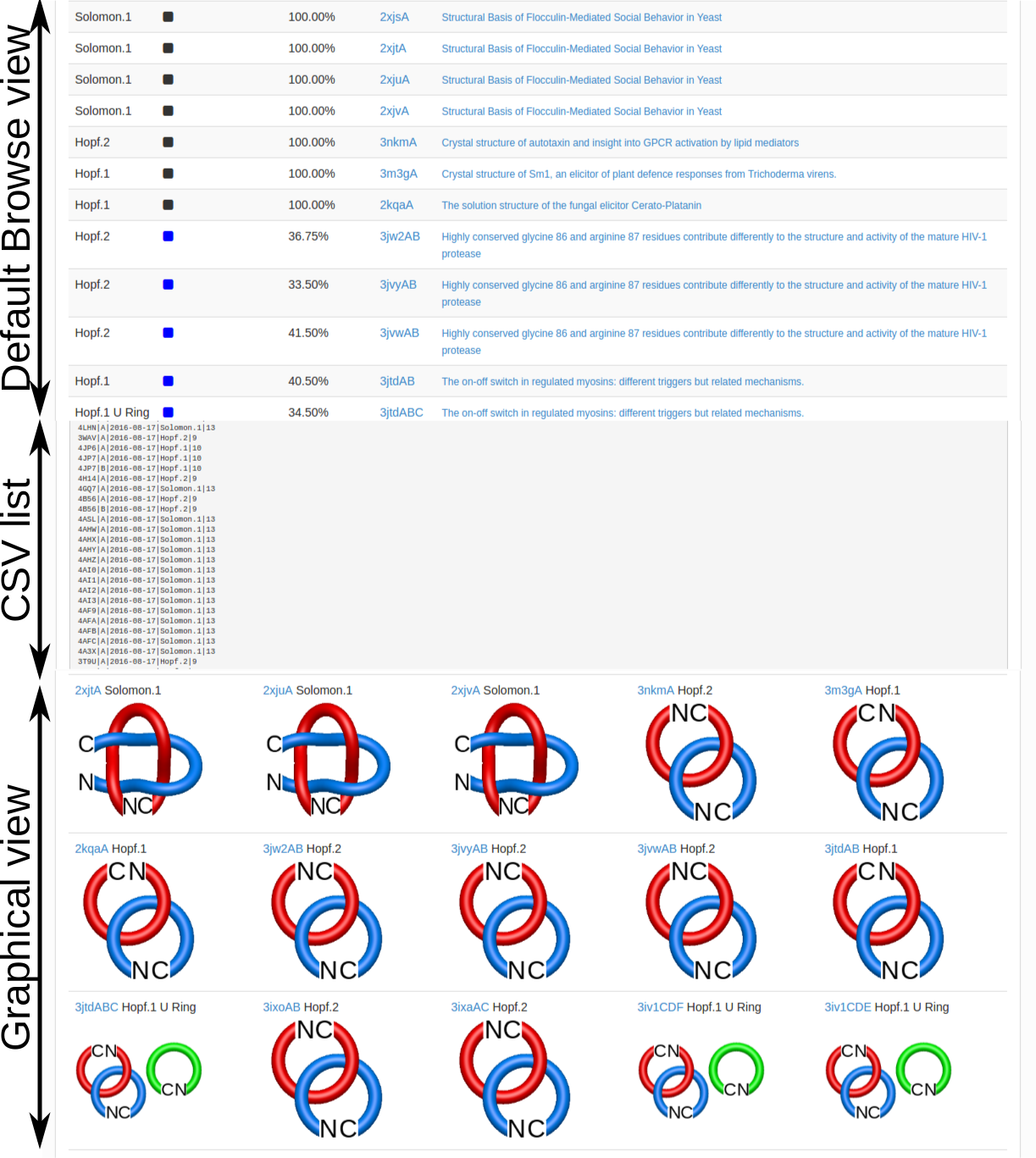
The user can also view the "raw data" in the form of CSV file for the table visible in default view of the database browse. This option can be useful for further computer-aided search through the database. The user can display the proteins with the schematic representation of corresponding topological motif (bottom panel in Fig. 1). This option is usefull for quick choice of the protein topology of interest.
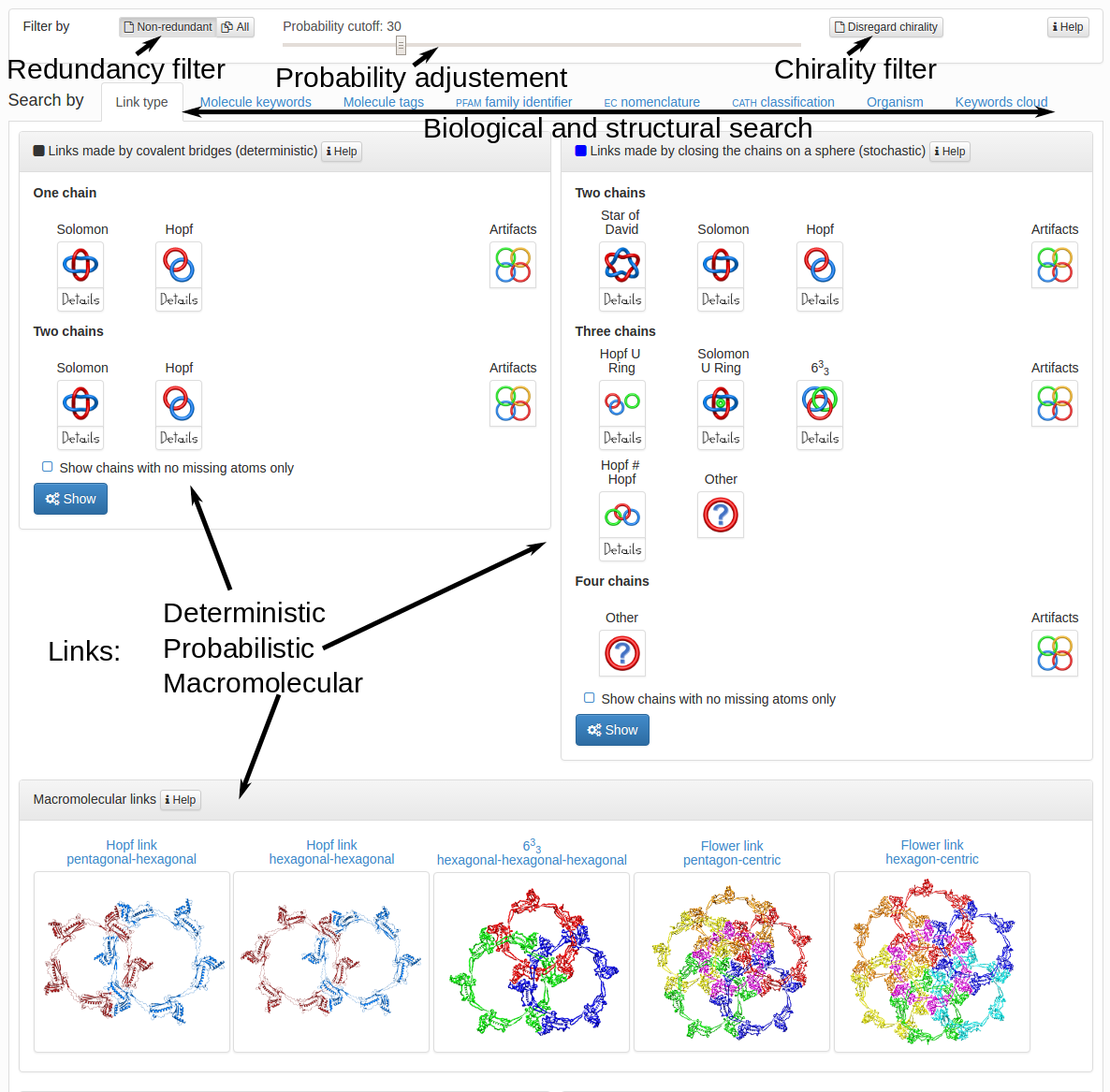
User can search the database in various ways. The main part of the search tab is divided into three parts corresponding to three types of links - deterministic (left-top frame), probabilistic (right-top frame) and macromolecular links (bottom frame) in Fig. 2. In each frame the user can choose wich structures should be displayed. After selecting topological types of interest, user has to click the "Show" button to display all of the structures matching the criteria choosen. Instead of selecting the major topological types (e.g. Hopf link), the user can select the structures with orientation or chirality specified (e.g. Hopf.1, see Fig. 3).
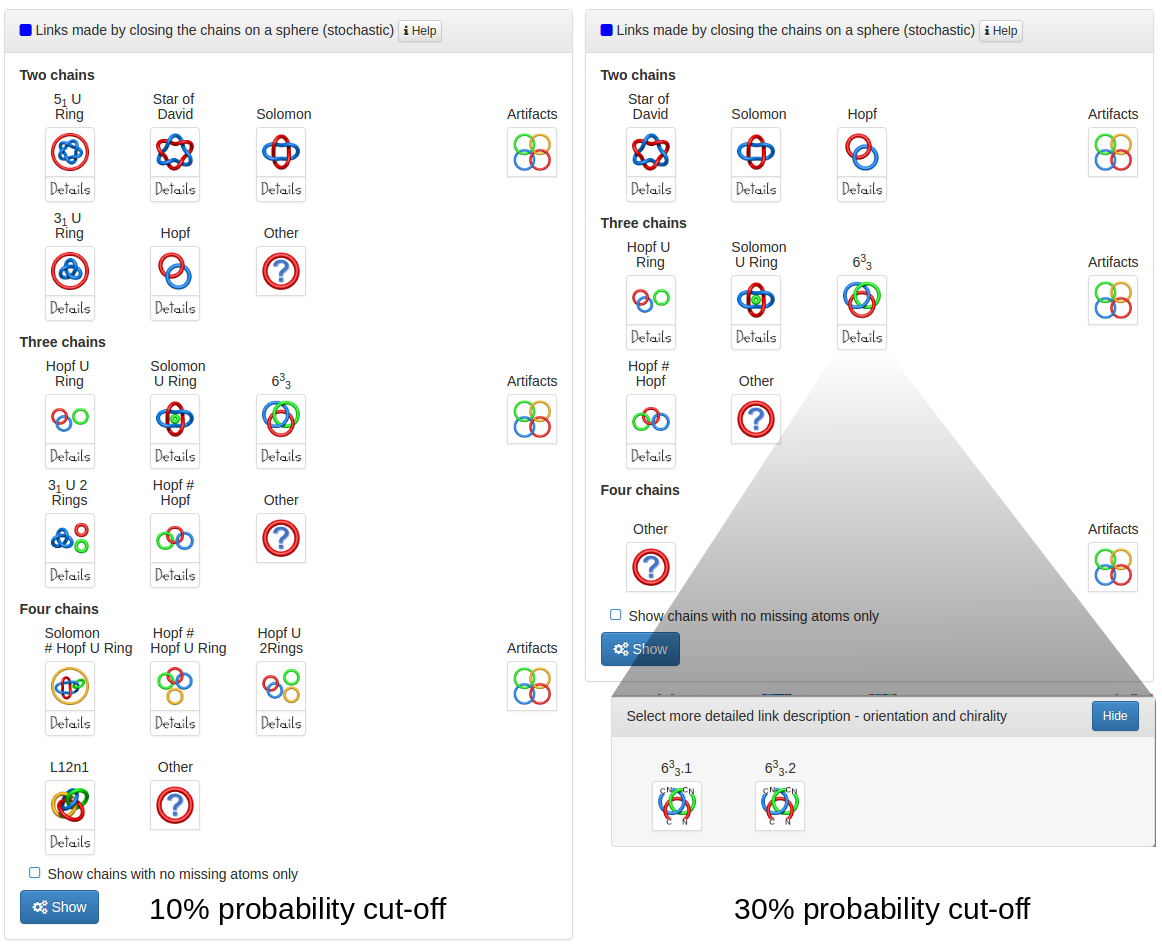
The content of the probabilistic links frame depends on the probability cut-off set in the top of the page. The default probability cut-off is set to 30% which means, that every protein with the likelihood of definite link topology higher than 30% will be displayed. Increasing the cut-off will decrease the number of proteins and topologies displayed (Fig. 3), but the links displayed will be more certain. One has to note however, that for a given cut-off the structures with chirality and orientation are displayed by default. It means, that if the cut-off is set to 40%, the structure with 25% of Hopf+ and 20% of Hopf- will not be displayed. To display the structures regardles of their chirality or orientation one has to click the "Disregard chirality" button in the top-right corner of the page. Activating this button will result in treating a structure with 25% of Hopf.1 and 20% of Hopf.2 as a structure with 45% of Hopf link likelihood.
There are two additional classes in deterministic and probabilistic links, i.e. Artifacts and Other. The artifact class include all the structures, which were found by our algorithm, but the existence of link is highly doubtfull. One of the reason the structure can be interpreted as an artifact is the occurence of very long gap. In the analysis, the gaps are joined by a straigth edge. If the gap is shorter or equal to 6 residues, introducing the straight interval instead of the piece of the chains should not change the topology. If however the gap is larger, the change of the topology is possible. Therefore the user has to browse the structure classified as the artifact with great caution. We plan to model the most interesting artifact structures in the future, to move them either to full links, or classify them as non-link structures.
The second special special class are the Other structures. It contains links, which topological type were not identified so far. The LinkProt database distinguishes all the links (either prime or composite) with 6 crossings and most prime links with 7 or 8 crossings. We try to identify succesfully all the unknown link with the algorith described in the link detection section.
The macromolecular links constitute separate class of links. They are divided according to the total topology and the number of chains forming each link component. Below the macromolecular links, there are the tables collecting the statistics of occurences of each individual class of links (except the macromolecular), including the orientation and chirality, separately for deterministic as well as for probabilistic links. Clicking on each class will result in printing the list of the links belonging to selected category. This is the alternative way to display the structures of interest. The statistics for the probabilistic links are also cut-off-dependent. By default, the structures shown in the main tab are the sequentially non-homologous structures. Therefore the numbers printed in the tables are applicable to statistics. However, to display all the structures the user can click on all button in the left-top corner of the page.