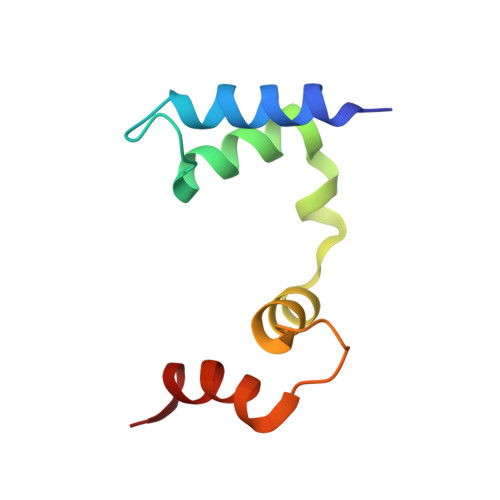
| Link type | Probability | Chain A piercings | Chain B piercings | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| view details |
|
Hopf.2 | 66% | +1B -75B -76B | -8A -22A +76A | ||||||||
| view details |
|
Hopf.2 | 66% | +1B -75B -76B | -8A -22A +76A | ||||||||
| view details |
|
Hopf.2 | 66% | +1B -75B -76B | -8A -22A +76A | ||||||||
| view details |
|
Hopf.2 | 66% | +1B -75B -76B | -8A -22A +76A | ||||||||
| view details |
|
Hopf.2 | 66% | +1B -75B -76B | -8A -22A +76A | ||||||||
| view details |
|
Hopf.2 | 66% | +1B -75B -76B | -8A -22A +76A | ||||||||
| view details |
|
Hopf.2 | 66% | +1B -75B -76B | -8A -22A +76A | ||||||||
Chain A Sequence |
MKSPEELKGIFEKYAAKEGDPNNLSKEELKLLLQTEFPSLLKGMSTLDELFEELDKNGDGEVSFEEFQVLVKKISQ |
Chain A Sequence |
MKSPEELKGIFEKYAAKEGDPNNLSKEELKLLLQTEFPSLLKGMSTLDELFEELDKNGDGEVSFEEFQVLVKKISQ |
| sequence length | 76,76 |
| structure length | 76,76 |
| publication title |
An extended hydrophobic core induces EF-hand swapping.
pubmed doi rcsb |
| molecule tags | Metal binding protein |
| molecule keywords | CALBINDIN D9K |
| source organism | Bos taurus |
| pdb deposition date | 2000-12-29 |
| LinkProt deposition date | 2016-10-21 |
| chain | Pfam Accession Code | Pfam Family Identifier | Pfam Description |
|---|---|---|---|
| AB | PF00036 | EF-hand_1 | EF hand |
| AB | PF01023 | S_100 | S-100/ICaBP type calcium binding domain |
| AB | PF00036 | EF-hand_1 | EF hand |
| AB | PF01023 | S_100 | S-100/ICaBP type calcium binding domain |
 Image from the rcsb pdb (www.rcsb.org)
Image from the rcsb pdb (www.rcsb.org)cath code
| Class | Architecture | Topology | Homology | Domain |
|---|---|---|---|---|---|
| Mainly Alpha | Orthogonal Bundle | Recoverin; domain 1 | EF-hand | ||
| Mainly Alpha | Orthogonal Bundle | Recoverin; domain 1 | EF-hand |
#chains in the LinkProt database with same CATH superfamily 1QX5 BKT; 1QX5 DI; 1QX5 DIRY; 3JTD AB; 1QX5 DIK; 2OPO AB; 1QX5 BIKT; 1QX7 ABDM; 2OPO AC; 1QX5 BIJK; 1QX7 IR; 1QX7 DIR; 1QX5 DJRT; 1QX5 JRY; 1QX5 JKTY; 1QX5 BIJR; 1ALV AB; 1QX5 DY; 1QX7 DIMR; 1QX5 DIT; 1QX5 BJY; 1QX5 BJTY; 1QX5 BDIY; 3JTD ABC; 1QX5 IJRY; 1QX5 JY; 1QX5 BIRY; 1QX5 KT; 1QX5 DIJY; 1QX7 AM; 1QX7 DI; 1QX7 AMR; 1QX5 DRY; 1QX5 JKRY; 1QX5 BDJT; 1QX5 IJK; 1QX5 BDJY; 1QX7 ABIM; 1QX7 ADM; 1QX5 IJKT; 1QX5 BK; 1QX5 BJK; 1QX5 BIJY; 1QX7 ABIR; 1QX7 ABDR; 1QX5 DR; 1QX5 BDIJ; 1QX7 ABMR; 1QX5 DIKR; 1QX5 BDI; 1A03 AB; 1QX5 DIRT; 1QX5 DKRT; 1QX5 DJY; 1QX5 BJKT; 1K9U AB; 1QX7 AD; 1QX7 IMR; 3JTD AC; 1QX5 IKT; 1QX5 DRTY; 1QX5 JKT; 1QX7 AIMR; 1QX5 DIJK; 1QX7 ADR; 1QX5 DIY; 2OPO BC; 2OPO ABCD; 1QX5 BIR; 1QX5 DJRY; 1QX5 BI; 1ALW AB; 1QX7 MR; 1QX7 DIM; 1QX5 BIK; 1QX7 AB; 1QX5 IKRT; 1QX5 BDIT; 1QX5 IKR; 1QX5 IKRY; 1QX5 BJ; 1HT9 AB; 1QX7 AIR; 1QX7 ADIR; 1QX5 DIJT; 1QX5 BDIK; 1QX7 ADI; 1QX7 ABI; 1QX5 DJKY; 1QX5 DRT; 1QX5 DJKT; 1QX5 IJKY; 1QX5 DTY; 1QX5 JRTY; 1QX5 DKTY; 1QX5 BIJT; 1QX7 ADIM; 1QX5 DT; 1QX7 ABD; 1QX5 DJT; 1QX5 JT; 2OPO ABC; 1QX5 IRY; 1QX5 DJTY; 1QX5 BIKR; 1QX5 IK; 1QX7 ABDI; 2OPO CD; 1QX5 JTY; 1QX5 IJKR; 1QX7 IM; 1QX5 JKY; 1QX5 BJRY; 1QX5 DKT; 1QX5 JK; 1QX7 DMR; 1QX5 DITY; 1QX5 RY; 1QX7 ABM; 1QX5 DIKY; 1QX5 BIJ; 1QX5 BDKT; 1QX7 ADMR; 1QX7 DR; 1QX5 BJT; 1QX5 BJKY; 1QX5 IR; 1QX5 DIR; 1QX7 AI; 1QX7 DM; 1QX5 DIKT; 2OPO BCD; 1QX7 AIM; 1QX5 BDIR; 2OPO ACD; #chains in the LinkProt database with same CATH topology 1QX5 BKT; 1QX5 DI; 1QX5 DIRY; 3JTD AB; 1QX5 DIK; 2OPO AB; 1QX5 BIKT; 1QX7 ABDM; 2OPO AC; 1QX5 BIJK; 1QX7 IR; 1QX7 DIR; 1QX5 DJRT; 1QX5 JRY; 1QX5 JKTY; 1QX5 BIJR; 1ALV AB; 1QX5 DY; 1QX7 DIMR; 1QX5 DIT; 1QX5 BJY; 1QX5 BJTY; 1QX5 BDIY; 3JTD ABC; 1QX5 IJRY; 1QX5 JY; 1QX5 BIRY; 1QX5 KT; 1QX5 DIJY; 1QX7 AM; 1QX7 DI; 1QX7 AMR; 1QX5 DRY; 1QX5 JKRY; 1QX5 BDJT; 1QX5 IJK; 1QX5 BDJY; 1QX7 ABIM; 1QX7 ADM; 1QX5 IJKT; 1QX5 BK; 1QX5 BJK; 1QX5 BIJY; 1QX7 ABIR; 1QX7 ABDR; 1QX5 DR; 1QX5 BDIJ; 1QX7 ABMR; 1QX5 DIKR; 1QX5 BDI; 1A03 AB; 1QX5 DIRT; 1QX5 DKRT; 1QX5 DJY; 1QX5 BJKT; 1K9U AB; 1QX7 AD; 1QX7 IMR; 3JTD AC; 1QX5 IKT; 1QX5 DRTY; 1QX5 JKT; 1QX7 AIMR; 1QX5 DIJK; 1QX7 ADR; 1QX5 DIY; 2OPO BC; 2OPO ABCD; 1QX5 BIR; 1QX5 DJRY; 1QX5 BI; 1ALW AB; 1QX7 MR; 1QX7 DIM; 1QX5 BIK; 1QX7 AB; 1QX5 IKRT; 1QX5 BDIT; 1QX5 IKR; 1QX5 IKRY; 1QX5 BJ; 1HT9 AB; 1QX7 AIR; 1QX7 ADIR; 1QX5 DIJT; 1QX5 BDIK; 1QX7 ADI; 1QX7 ABI; 1QX5 DJKY; 1QX5 DRT; 1QX5 DJKT; 1QX5 IJKY; 1QX5 DTY; 1QX5 JRTY; 1QX5 DKTY; 1QX5 BIJT; 1QX7 ADIM; 1QX5 DT; 1QX7 ABD; 1QX5 DJT; 1QX5 JT; 2OPO ABC; 1QX5 IRY; 1QX5 DJTY; 1QX5 BIKR; 1QX5 IK; 1QX7 ABDI; 2OPO CD; 1QX5 JTY; 1QX5 IJKR; 1QX7 IM; 1QX5 JKY; 1QX5 BJRY; 1QX5 DKT; 1QX5 JK; 1QX7 DMR; 1QX5 DITY; 1QX5 RY; 1QX7 ABM; 1QX5 DIKY; 1QX5 BIJ; 1QX5 BDKT; 1QX7 ADMR; 1QX7 DR; 1QX5 BJT; 1QX5 BJKY; 1QX5 IR; 1QX5 DIR; 1QX7 AI; 1QX7 DM; 1QX5 DIKT; 2OPO BCD; 1QX7 AIM; 1QX5 BDIR; 2OPO ACD; #chains in the LinkProt database with same CATH homology 1QX5 BKT; 1QX5 DI; 1QX5 DIRY; 3JTD AB; 1QX5 DIK; 2OPO AB; 1QX5 BIKT; 1QX7 ABDM; 2OPO AC; 1QX5 BIJK; 1QX7 IR; 1QX7 DIR; 1QX5 DJRT; 1QX5 JRY; 1QX5 JKTY; 1QX5 BIJR; 1ALV AB; 1QX5 DY; 1QX7 DIMR; 1QX5 DIT; 1QX5 BJY; 1QX5 BJTY; 1QX5 BDIY; 3JTD ABC; 1QX5 IJRY; 1QX5 JY; 1QX5 BIRY; 1QX5 KT; 1QX5 DIJY; 1QX7 AM; 1QX7 DI; 1QX7 AMR; 1QX5 DRY; 1QX5 JKRY; 1QX5 BDJT; 1QX5 IJK; 1QX5 BDJY; 1QX7 ABIM; 1QX7 ADM; 1QX5 IJKT; 1QX5 BK; 1QX5 BJK; 1QX5 BIJY; 1QX7 ABIR; 1QX7 ABDR; 1QX5 DR; 1QX5 BDIJ; 1QX7 ABMR; 1QX5 DIKR; 1QX5 BDI; 1A03 AB; 1QX5 DIRT; 1QX5 DKRT; 1QX5 DJY; 1QX5 BJKT; 1K9U AB; 1QX7 AD; 1QX7 IMR; 3JTD AC; 1QX5 IKT; 1QX5 DRTY; 1QX5 JKT; 1QX7 AIMR; 1QX5 DIJK; 1QX7 ADR; 1QX5 DIY; 2OPO BC; 2OPO ABCD; 1QX5 BIR; 1QX5 DJRY; 1QX5 BI; 1ALW AB; 1QX7 MR; 1QX7 DIM; 1QX5 BIK; 1QX7 AB; 1QX5 IKRT; 1QX5 BDIT; 1QX5 IKR; 1QX5 IKRY; 1QX5 BJ; 1HT9 AB; 1QX7 AIR; 1QX7 ADIR; 1QX5 DIJT; 1QX5 BDIK; 1QX7 ADI; 1QX7 ABI; 1QX5 DJKY; 1QX5 DRT; 1QX5 DJKT; 1QX5 IJKY; 1QX5 DTY; 1QX5 JRTY; 1QX5 DKTY; 1QX5 BIJT; 1QX7 ADIM; 1QX5 DT; 1QX7 ABD; 1QX5 DJT; 1QX5 JT; 2OPO ABC; 1QX5 IRY; 1QX5 DJTY; 1QX5 BIKR; 1QX5 IK; 1QX7 ABDI; 2OPO CD; 1QX5 JTY; 1QX5 IJKR; 1QX7 IM; 1QX5 JKY; 1QX5 BJRY; 1QX5 DKT; 1QX5 JK; 1QX7 DMR; 1QX5 DITY; 1QX5 RY; 1QX7 ABM; 1QX5 DIKY; 1QX5 BIJ; 1QX5 BDKT; 1QX7 ADMR; 1QX7 DR; 1QX5 BJT; 1QX5 BJKY; 1QX5 IR; 1QX5 DIR; 1QX7 AI; 1QX7 DM; 1QX5 DIKT; 2OPO BCD; 1QX7 AIM; 1QX5 BDIR; 2OPO ACD;
#similar chains in the LinkProt database (?% sequence similarity) ...loading similar chains, please wait... #similar chains, but unlinked ...loading similar chains, please wait... #similar chains in the pdb database (?% sequence similarity) ...loading similar chains, please wait...